Part II. Future Shock and Selenocysteine: it's time again to update the databanks
One of the surprises (for me anyway) in discovering the existence of selenocysteine (Part I. Future Shock and Selenocysteine), was the corresponding discovery that it's encoded by UGA. Ordinarily, UGA is a stop codon. If a UGA is in an mRNA sequence, it tells the ribosome that the job is done. It's time to pack up all the tRNAs and elongation factors and move on to the next project.
But in the case of selenocysteine, we have a "work-around." Sometimes UGA stops everything, sometimes the UGA says "put the selenocysteine right here." (Someone in the office joked that the ID g-o-d must be a programmer since he/she is trying to fix bugs.)
How do the ribosomes and tRNAs know whether to stop or go?
They feel the difference.
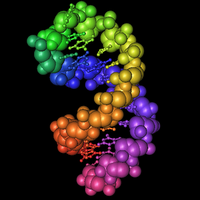 Seriously, the sequences at the 3' end of an mRNA fold into a special hairpin shape like the one shown here (1, 2, 3). In bacteria, this structure is called a "selenocysteine insertion sequence" or SECIS element. Eucaryotes have similar structures at the 3' ends of mRNAs for selenoproteins.
Seriously, the sequences at the 3' end of an mRNA fold into a special hairpin shape like the one shown here (1, 2, 3). In bacteria, this structure is called a "selenocysteine insertion sequence" or SECIS element. Eucaryotes have similar structures at the 3' ends of mRNAs for selenoproteins.
The RNA in the picture has a rainbow coloring scheme (Red, Orange, Yellow, Green, Blue, Indigo, Violet). Nucleotides at the 5' end are red, nucleotides at the 3' end are violet. You can follow the colors in the RNA backbone to see how the RNA is twisted around into a hairpin shape.
One of the tests that we used to give job applicants was to have them write a short script for translating a DNA sequence in 6 reading frames. We would give them a mouse pad with the genetic code and put them to work.
Selenocysteine makes this problem a whole lot harder.
Since the recognition feature is a secondary structure, locating the coding sequences for selenocysteines presents an interesting challenge to computational biologists. Finding these sequences requires a bit more than a regular expression.
Kryukov, et. al. describe an algorithm for doing this type of search (1). They've refined it in the years since this publication, but it seems that the information has yet to percolate through much of the world's bioinformatics community.
And here, I thought I was the only one who seemed to have missed this.
Nope.
Can we find selenocysteine in GenBank?
I started to wonder if the news about selenocysteine had trickled out beyond PubMed articles and into the rest of the NCBI.
Could I find selenoprotein sequences in the Gene database? I thought this would be a good place to start since the data are well curated and there are links to reference protein sequences.
I searched and searched, and lo and behold, I found them.

The sequence above codes for human selenoprotein P. U is the one letter symbol that represents selenocysteine. This protein contains an unusually large number of selenocysteines.
I only looked at a few of the reference protein sequences (labeled NP---) from the Gene database, but they all seemed to have selenocysteines.
So the NCBI Gene Database seems to be caught up, at least for the sequences that I checked out.
Mischief and Misannotations
I followed the links to the Conserved Domain Database. (I'm writing a book on this BTW, and the CDD is really, really cool).
When I got to a summary page, I choose the SelP_C domain (since more U's are on that side of the protein). This gave me a page with a Pfam alignment between my human SelP sequence and some sequences that were chosen for Pfam. (you can take a look at this yourself by clicking the link above. Change the format to Hypertext and click Show Alignment to see the selenocysteines in the query sequence).
Reading downward, the Pfam sequences, in the alignment below, are from cow, my query(human), rat, another human sequence, and zebrafish.

Every time my query sequence has a "U," the other sequences have a "c" (purple boxes above).
This is interesting and odd. Only one of the proteins with the conserved SelP domain has selenocysteine (and it's our human query sequence).
One interpretation that Kryukov suggested in 2003 (1), (and later regretted, I'm sure), is that through evolution, cysteine was substituted for selenocysteine in organisms like the rat and mouse.
I think the presence of the other human SelP sequence argues for another interpretation, especially since it's an older version of our query.
If we click the gi links to see the database records, we find something else that's interesting.
A note in the first sequence, from the cow, deposited in April 2006, shows that someone knew about the selenocysteines,
Maybe they didn't read the note.
Stranger, yet, the missing selenocysteines could be rationalized away by arguing that the protein sequence is just a conceptual translation - that is, it was determined by using the standard genetic code. Except that using the standard genetic code would have generated a much shorter sequence since UGA makes translation stop. So, instead of putting in the correct amino acid, the curators (Swiss prot?) typed in the wrong amino acid. Instead of using the U for selenocysteine, they typed a C for cysteine.
The rat sequence was also updated in April 2006 and we can see that the positions of selenocysteines also seem to be marked in the GenPept record (below), but, funny, there aren't any selenocysteines in the rat sequence.

The other human sequence for SelP and the zebrafish sequence show the same kinds of annotations. Yet neither one contains selenocysteines in the amino acid sequence.
Could the source of the sequences (Pfam) be the source of the problems?
I'm not sure where the problem originates but if I search the Pfam database at the Sanger Center, for selenocysteine, I get a list of 31 proteins that contain it, and again, I get an annotation that indicates that someone is aware of selenocysteine.
If I take this sequence and do a blastp search at the NCBI, I get quite few perfect matches. Just like Pfam, there are sequences in GenBank that are not yet fixed.
What is our take home message?
The simple take-home message, of course, is to be aware the FASTA sequences for selenium-containing proteins are likely to be wrong. If the annotations say there should be selenocysteine and you can't find a "u" in the sequence, it probably hasn't been fixed yet. Those of us who use the date must always be skeptical and read the literature.
The second take-home message concerns process. The acceptance of new ideas in science generally prompts some re-evaluation of older ideas. We evaluate older concepts more critically in the electric light of new ideas. It would be helpful if these processes could be applied more quickly to sequence data and bioinformatics algorithms. These results support the need for scientific curators who can read the literature, add annotations, and even make corrections in amino acid sequences, from time to time.
The amino acid matrices that we use for protein comparisons, were updated when more sequences became available for doing alignments. We all switched from using PAM to BLOSUM matrices. Maybe it's time to make update the Pfam domains as well.
Selenocysteine exists.
It's time to deal with it and get on with the work.
References:
1. Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. 2003. Characterization of mammalian selenoproteomes.
Science. 300:1439-43.
2. Diamond, AM. 2004. On the road to selenocysteine. Proc Natl Acad Sci U S A. 101: 13395-13396.
3. Yoshizawa S, Rasubala L, Ose T, Kohda D, Fourmy D, Maenaka K. 2005. Structural basis for mRNA recognition by elongation factor SelB. Nat Struct Mol Biol. 12:198-203.
Subject: Doing biology with bioinformatics
But in the case of selenocysteine, we have a "work-around." Sometimes UGA stops everything, sometimes the UGA says "put the selenocysteine right here." (Someone in the office joked that the ID g-o-d must be a programmer since he/she is trying to fix bugs.)
How do the ribosomes and tRNAs know whether to stop or go?
They feel the difference.
 Seriously, the sequences at the 3' end of an mRNA fold into a special hairpin shape like the one shown here (1, 2, 3). In bacteria, this structure is called a "selenocysteine insertion sequence" or SECIS element. Eucaryotes have similar structures at the 3' ends of mRNAs for selenoproteins.
Seriously, the sequences at the 3' end of an mRNA fold into a special hairpin shape like the one shown here (1, 2, 3). In bacteria, this structure is called a "selenocysteine insertion sequence" or SECIS element. Eucaryotes have similar structures at the 3' ends of mRNAs for selenoproteins.The RNA in the picture has a rainbow coloring scheme (Red, Orange, Yellow, Green, Blue, Indigo, Violet). Nucleotides at the 5' end are red, nucleotides at the 3' end are violet. You can follow the colors in the RNA backbone to see how the RNA is twisted around into a hairpin shape.
One of the tests that we used to give job applicants was to have them write a short script for translating a DNA sequence in 6 reading frames. We would give them a mouse pad with the genetic code and put them to work.
Selenocysteine makes this problem a whole lot harder.
Since the recognition feature is a secondary structure, locating the coding sequences for selenocysteines presents an interesting challenge to computational biologists. Finding these sequences requires a bit more than a regular expression.
Kryukov, et. al. describe an algorithm for doing this type of search (1). They've refined it in the years since this publication, but it seems that the information has yet to percolate through much of the world's bioinformatics community.
And here, I thought I was the only one who seemed to have missed this.
Nope.
Can we find selenocysteine in GenBank?
I started to wonder if the news about selenocysteine had trickled out beyond PubMed articles and into the rest of the NCBI.
Could I find selenoprotein sequences in the Gene database? I thought this would be a good place to start since the data are well curated and there are links to reference protein sequences.
I searched and searched, and lo and behold, I found them.

The sequence above codes for human selenoprotein P. U is the one letter symbol that represents selenocysteine. This protein contains an unusually large number of selenocysteines.
I only looked at a few of the reference protein sequences (labeled NP---) from the Gene database, but they all seemed to have selenocysteines.
So the NCBI Gene Database seems to be caught up, at least for the sequences that I checked out.
Mischief and Misannotations
I followed the links to the Conserved Domain Database. (I'm writing a book on this BTW, and the CDD is really, really cool).
When I got to a summary page, I choose the SelP_C domain (since more U's are on that side of the protein). This gave me a page with a Pfam alignment between my human SelP sequence and some sequences that were chosen for Pfam. (you can take a look at this yourself by clicking the link above. Change the format to Hypertext and click Show Alignment to see the selenocysteines in the query sequence).
Reading downward, the Pfam sequences, in the alignment below, are from cow, my query(human), rat, another human sequence, and zebrafish.

Every time my query sequence has a "U," the other sequences have a "c" (purple boxes above).
This is interesting and odd. Only one of the proteins with the conserved SelP domain has selenocysteine (and it's our human query sequence).
One interpretation that Kryukov suggested in 2003 (1), (and later regretted, I'm sure), is that through evolution, cysteine was substituted for selenocysteine in organisms like the rat and mouse.
I think the presence of the other human SelP sequence argues for another interpretation, especially since it's an older version of our query.
If we click the gi links to see the database records, we find something else that's interesting.
A note in the first sequence, from the cow, deposited in April 2006, shows that someone knew about the selenocysteines,
[MISCELLANEOUS] The selenocysteines are all encoded by the opal codon, UGA.But apparently, no one bothered to put them in the amino acid sequence, since there aren't any selenocysteines there.
Maybe they didn't read the note.
Stranger, yet, the missing selenocysteines could be rationalized away by arguing that the protein sequence is just a conceptual translation - that is, it was determined by using the standard genetic code. Except that using the standard genetic code would have generated a much shorter sequence since UGA makes translation stop. So, instead of putting in the correct amino acid, the curators (Swiss prot?) typed in the wrong amino acid. Instead of using the U for selenocysteine, they typed a C for cysteine.
The rat sequence was also updated in April 2006 and we can see that the positions of selenocysteines also seem to be marked in the GenPept record (below), but, funny, there aren't any selenocysteines in the rat sequence.

The other human sequence for SelP and the zebrafish sequence show the same kinds of annotations. Yet neither one contains selenocysteines in the amino acid sequence.
Could the source of the sequences (Pfam) be the source of the problems?
I'm not sure where the problem originates but if I search the Pfam database at the Sanger Center, for selenocysteine, I get a list of 31 proteins that contain it, and again, I get an annotation that indicates that someone is aware of selenocysteine.
SelP is the only known eukaryotic selenoprotein that contains multiple selenocysteine (Sec) residues...Yet when I do a seed alignment, none of the amino acid sequences contain selenocysteine. Here is one example:
SEPP1_HUMAN/22-250 QDQSSLCKQPPAWSIRDQDPMLNSNGSVTVVALLQASCYLCILThe selenocysteines are missing here, too.
QASKLEDLRVKLKKEGYSNISYIVVNHQGISSRLKYT
If I take this sequence and do a blastp search at the NCBI, I get quite few perfect matches. Just like Pfam, there are sequences in GenBank that are not yet fixed.
What is our take home message?
The simple take-home message, of course, is to be aware the FASTA sequences for selenium-containing proteins are likely to be wrong. If the annotations say there should be selenocysteine and you can't find a "u" in the sequence, it probably hasn't been fixed yet. Those of us who use the date must always be skeptical and read the literature.
The second take-home message concerns process. The acceptance of new ideas in science generally prompts some re-evaluation of older ideas. We evaluate older concepts more critically in the electric light of new ideas. It would be helpful if these processes could be applied more quickly to sequence data and bioinformatics algorithms. These results support the need for scientific curators who can read the literature, add annotations, and even make corrections in amino acid sequences, from time to time.
The amino acid matrices that we use for protein comparisons, were updated when more sequences became available for doing alignments. We all switched from using PAM to BLOSUM matrices. Maybe it's time to make update the Pfam domains as well.
Selenocysteine exists.
It's time to deal with it and get on with the work.
References:
1. Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. 2003. Characterization of mammalian selenoproteomes.
Science. 300:1439-43.
2. Diamond, AM. 2004. On the road to selenocysteine. Proc Natl Acad Sci U S A. 101: 13395-13396.
3. Yoshizawa S, Rasubala L, Ose T, Kohda D, Fourmy D, Maenaka K. 2005. Structural basis for mRNA recognition by elongation factor SelB. Nat Struct Mol Biol. 12:198-203.
Subject: Doing biology with bioinformatics
technorati tags: bioinformatics, selenocysteine, biochemistry, biology, blast, genetics, genomics, DNA, RNA, Science Education

16 Comments:
Check this out. UGA is actually over represented eukaryotes among termination codons. I would have expected that it would under represented so as to prevent accidental non-termination. I couldn't find anything on prokaryotes. Is most of the work on selenocysteine from bacteria? If so, the usage pattern of stop codons may tell a different story.
Selenocysteines are found in bacteria, eucaryotes, and archeabacteria. If I go the PubMed reference for the first citation and click the link to get papers by Kryukov, I can find papers on Plasmodium, nematodes, bacteria, Chlamydomonas, mammals, fruit flies, algae.
In this Jan 2006 paper, the authors state in the first sentence that selenoproteins are only found in animals and algae, then they go on to find selenoproteins in the Plasmodium genome that were there, but misannotated.
My impression is that a global search of the taxonomic kingdom is hard to do, given the issues with annotation. I don't know how hard it is to run their program and look for SECIS elements, but it seems that something like this would be worthwhile.
Wow!
Great! Yet another discovery that makes biology so exciting. We never have to worry about teaching the same thing year in and year out. I need to check this stuff out for myself...mmm time for a BLAST!
Now you've done it. I just squandered a perfectly good evening on NCBI. Actually did get lots of interesting stuff about selenoproteins but it is going to take me some time to digest it. On a related note have you seen this article from Scientific American?
Scientific American: Evolution Encoded [ BIOTECHNOLOGY ]New discoveries about the rules governing how genes encode proteins have revealed nature's sophisticated "programming" for protecting life from catastrophic errors while accelerating evolution
If selenocysteine worries you, try pyrollysine. It doesn't even have a one-letter code
assigned to it yet. We couldn't represent it in a standard way even if we wanted to.
Most comp bio software works with a 20-letter basic aa code (rather than 22 for known genetically encoded aa's, and zillions if you include posttranslational modification), because it's a reasonable approximation, given the rarity of selC and pyrrolysine.
So yes, we know about selC. (I'm the author of the HMMER software that Pfam is built with. HMMER uses a base 20 letter alphabet, so it's forced to represent selenocysteine as something - it chooses C semi-arbitrarily.) Ignoring it is a design decision, and it's not the only design decision that comp bio software makes in approximating biology efficiently.
Hi Anon,
Thanks for the clearing up the info on Pfam and the heads up on prolyl lysine (yikes!).
I do understand the logic.
The problem though, is that there's no way for a biologist to know if a couple of amino acids were left out because of a design decision or if they just don't occur in that protein.
How are we know when the public databases and processed data really mimick the real world?
And, of course, we biologists have a tendency to get fixated on the unusual and rare situations. :- )
I have seen our perceptions change as more data are made available. When I first started sequencing DNA, alternative splicing was considered rare and unusual. Not anymore.
As we do more and more proteomics and increase the volume of structural data, we will probably find lots of rare modifications. An interesting challenge will be how to modify software and data structures to match the new things we learn.
"SelP is the only known eukaryotic selenoprotein that contains multiple selenocysteine (Sec) residues..."
That's not 100% true, since deiodinase type 2 also can translate an UGA located close to the stop UGA into selenocysteine.
There are some bioinformatitians already working on solving the kind of problems you suggest. There's a software that recognizes potential SECIS elements on a sequence, and that's described here.
Selenoprotein function is usually related to redox and cell detoxification from oxidative stress, but they also control thyroid hormone metabolism, for example.
And to spice your curiousity up, there are selenoproteins in Archaea, in bacteria, in eukaryotes, but none in yeasts.
(A last tip: I think you crossed by my last paper while literature searching for this Tangled Bank edition...)
;-)
Good job.
Thanks!!
I appreciate the update and the quick synopsis of selenoprotein function.
Are you sure that yeast don't have selenoproteins? I don't believe we can rely on databases for this kind of info. I think we need some good analytical chemistry and some mass spec data.
Anyway, I'll go back and read the paper a bit more carefully. ; - )
In the case of yeast, research was not only done with databases, but also with radioactive labeling with Se75. The evidence that yeast lacks selenoproteins rely on a number of functional experiments, obtained prior to the big boom of bioinformatics in the 90's. But they do have homologues with cysteine instead of selenocysteine.
Thanks!
That sounds pretty conclusive. I can't think of any experiments you could do to top that.
What about other fungi?
Slime mold contains cysteine-homologues too. There's no information in the literature about other fungi studies. I would imagine that people have tried hard to find it, but unfortunately "negative" results not often make through peer-review... ;-)
This paper has a neat overview table of all selenocysteine/cysteine genes found and its respective groups.
Thanks for the clearing up the info on Pfam and the heads up on prolyl lysine (yikes!)
Just a little more clearing-up; it really is pyrrolysine. Wikipedia link. I think "pyrro" refers to the cyclisation around the N (see structure).
The original paper is referenced here.
As anonymous (hi Sean!) mentioned, one problem in computational biology is that a lot of software expects the "standard" 20 amino acid symbols as input and will complain otherwise. As for databases, I remember my initial confusion on encountering "U" in sequences for the first time some years ago. This kind of thing is probably best represented not by extending the alphabet but by using controlled vocabularies, structured data and better curation in databases. Things like UniProt will have comment lines that mention unusual features.
Another problem arises when you want to annotate genes in a new genome sequence. Current gene-finding software will treat an amber codon simply as a stop codon, so if it's really an amber codon, you'll get 2 open reading frames (in simple cases like prokaryotes). If the protein is known of course, then they'll both give a BLAST hit to the same sequence, which will give you a clue. We had this situation in an archaeon that I worked on - when we came to do proteomics and identified spots from a 2D gel, we found one spot mapping to two neighbouring open reading frames - closer inspection showed that we were looking at read-through and most likely, pyrrolysine. There's some software around that screens for the selenocysteine case, but pyrrolysine incorporation seems to work differently.
Always plenty of new challenges in bioinformatics :)
Thanks Selenoprotein researcher and thanks Neil!
(Don't you guys ever sleep?)
I never expected to learn so much from the comment section!
I got a cool SeC story for ya.
The deiodinase 2 gene was implicated in a QTL analysis I did for my graduate work, so I learned all about SECIS elements and coding UGAs, since SeC is vital to the function of Dio2. Therefore it made sense to sequence not just the coding region of the gene when looking for alterations, but the 3' UTR, which I did. There's an unusually long stretch between the Dio2 coding region and its SECIS element--something like 5 kb, when most other SECISes are a few hundred bp away-- such that no mRNA containing both had been isolated. I had to ID the SECIS directly from genomic sequence in silico to design primers that went out far enough.
The then-newly-available genome browsers found something about Dio2 that had gone unnoticed until then. A gene called PORF1 falls in between Dio2 and its predicted SECIS element. It's one of those genes that is the life work of one person and nobody else has ever heard of it, yet there it is in the databases anyway. PORF1 is itself a selenoprotein, several times over, even (IIRC it has 2 UGA and maybe more). What's more, the northern blots identifying it showed RNAs of a length consistent with the whole shebang being expressed in one long transcript (Dio2, PORF1, SECIS). It also showed a lot of other existing lengths of the mRNA. Add it all together and: Dio2 (which regulates levels of thyroid hormone in the brain and so is therefore a leetle bit important) is competing with another transcript for use of the SECIS element they share.
And lastly....guess what PORF1 encodes, when the UGAs are retranslated? A transcription factor. The work hasn't been done on what it regulates, but since it competes with Dio2 for the SECIS, the more PORF1 made, the less Dio2 there is. Doesn't it sound like PORF1 should be a positive regulator of Dio2/negative regulator of itself? In this way the levels of SeC can directly regulate Dio2 and therefore thyroid hormone levels.
Now the reason I could never get anyone else interested in this story is that it's not true in the human Dio2, whose 3' UTR doesn't have a PORF-like ORF. But I still maintain that it's interesting from the genomics perspective as a novel method of regulation. The alternative splicing example is a good one to remember; we thought it was soooo weird and now it appears to be the norm...how many more cases like that, or like PORF1/Dio2, are there out there??
I'm currently doing whole-genome association studies, and clearly we have no fricking idea about how regulation happens. We keep looking for non-synonymous SNPs only because we can't make a case for how other ones could be working. My favorite metaphor for this: a person is looking for something at night under a streetlight. An observer asks, "What're you looking for?" "My keys." "Where'd you lose them?" "Over there" (points to dark area). "Why the heck aren't you looking over there?" "The light's better here."
(PORF/DIO2 ref available upon request.)
Wow!! Can you send me (or post) the reference? I would like to read it.
This has turned into a more interesting story than (I ever anticipated.
This is a great discussion about Selenocysteine.
Post a Comment
<< Home